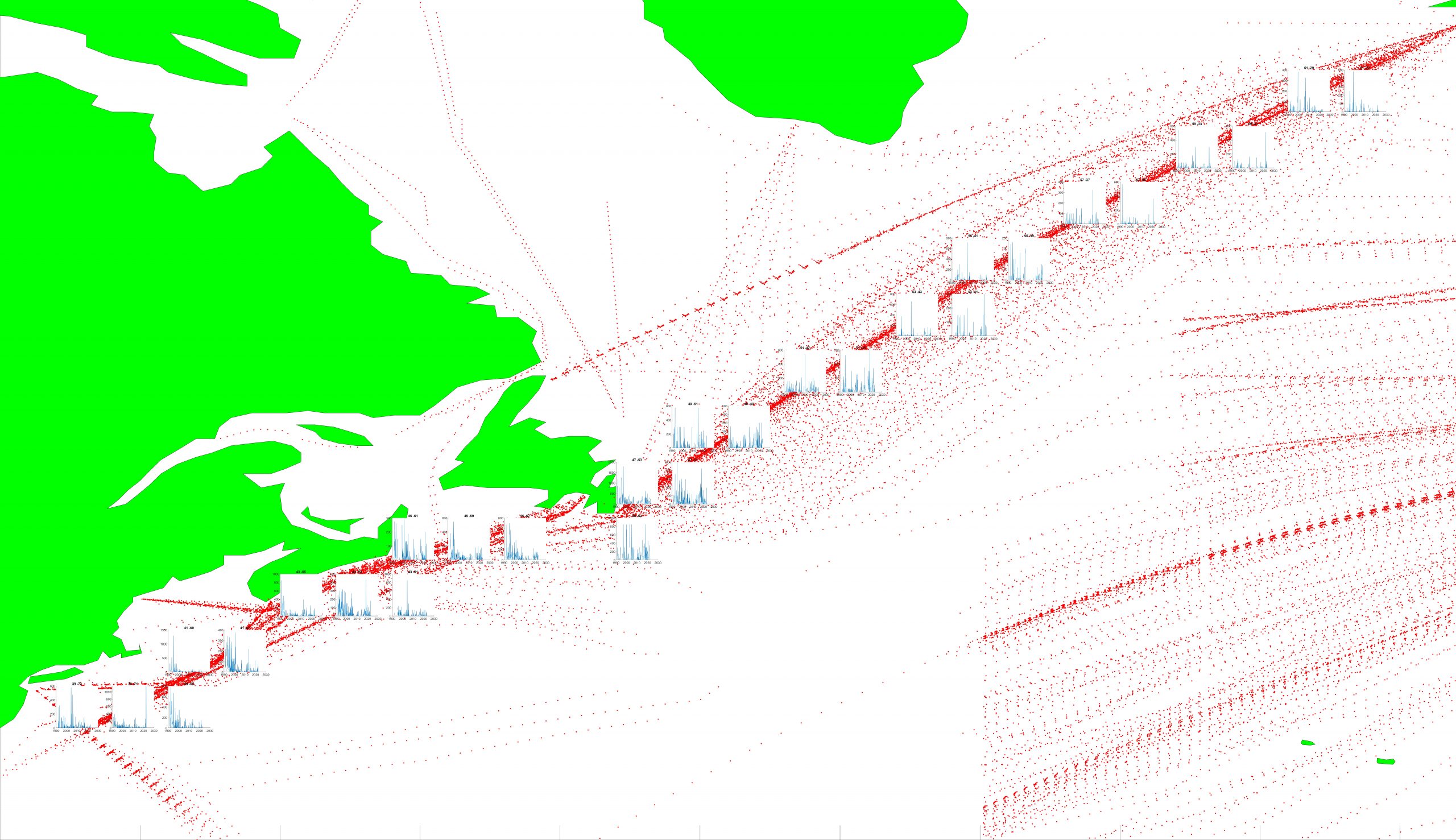
Data Methodology
Here we present Matlab scripts to extract samples and construct time-series for common zooplankton and phytoplankton taxa in the CPR-BEAMS dataset
Matlab scripts
Scripts to Extract Data and Generate Time-Series
The scripts are available via GitHub:
lws210725/CPR-BEAMS: Example MATLAB scripts for working with CPR-BEAMS data via BCO-DMO
Functions of the Scripts:
Data Reader with Area Restriction
- Extract and map the locations of available samples within a specified area and range of dates from the CPR-BEAMS resource on BCO-DMO.
- Extract abundance data for specified taxa from the CPR-BEAMS resource on BCO-DMO.
- Return a monthly time-series for the abundance of a specified taxon within an example area and range of dates:
- Highlight missing samples.
- Backfill missing data using the local median value for any given month.
Data Reader and Gridder
- Grid the data and produce localized seasonal abundance profiles for representative example taxa.
- Grid the data and produce localized monthly abundance time-series for representative example taxa.
- Grid the data and produce localized monthly abundance time-series for representative example taxa:
- Highlight missing samples.
- Backfill missing data using the local median value for any given month.
Contact:
For questions, clarifications, or assistance with the scripts or the CPR-BEAMS data resource, please contact Lawrence Sheppard. He can provide support on:
- Understanding how to use the scripts to extract and analyze data.
- Troubleshooting issues with data extraction, mapping, or time-series generation.
- Customizing the scripts for specific research needs or datasets.